Research Intern at IIT-Bombay

During my research internship at IIT Bombay under the guidance of Professor Kantimay Das Gupta, I had the opportunity to work on an innovative and impactful project focused on the "Development of a PNA–DiSc 2 Based Portable Absorbance Platform for the Detection of Pathogen Nucleic Acids." This project was at the intersection of molecular biology, bioengineering, and computational systems, aimed at developing a cost-effective, rapid, and portable diagnostic tool for detecting pathogen-specific nucleic acids. The primary goal of the project was to design and optimize a platform that leverages Peptide Nucleic Acid (PNA) probes and a novel dye, DiSc 2, to enable the highly sensitive and specific detection of nucleic acids. PNAs are synthetic analogs of DNA with a neutral backbone, which grants them enhanced stability and specificity in hybridization reactions. The use of DiSc 2, a dye sensitive to changes in absorbance, provided a robust mechanism for signal transduction upon target binding. This combination was instrumental in ensuring the platform's reliability for pathogen detection.
- Developed the platform's software using Python, leveraging libraries like PyQt for the touch display user interface
- Designed and implemented mathematical algorithms for absorbance data analysis and pathogen detection
- Used NumPy, Tkinter and SciPy for computational calculations and data processing to ensure precise and efficient performance.
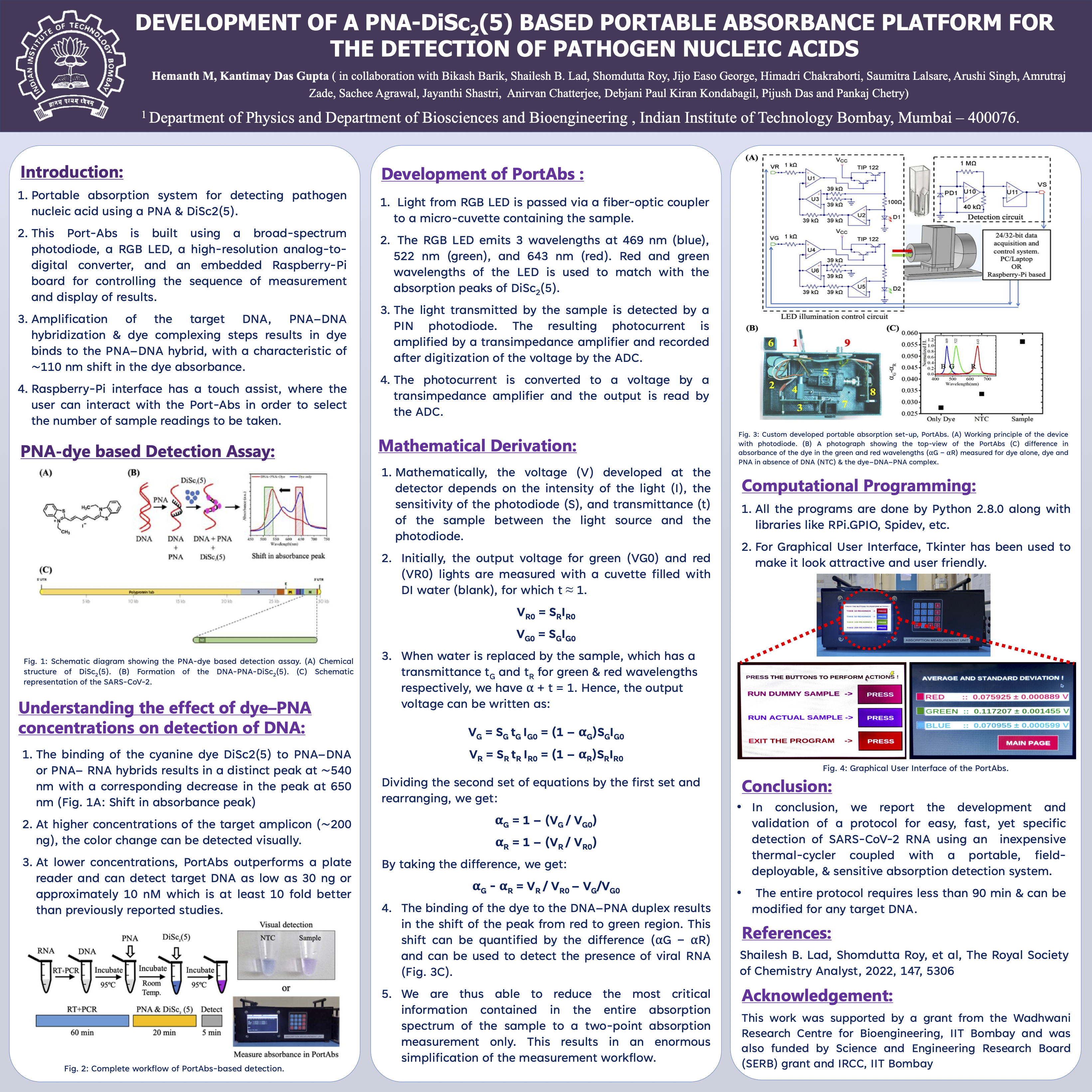
My primary responsibility in the project was to code the entire system and develop a user interface in Python, designed for a touch display. This involved writing the software to process absorbance data, perform mathematical calculations, and control the hardware components of the platform. I implemented algorithms to analyze and interpret absorbance patterns, setting thresholds for pathogen detection. Additionally, I ensured seamless integration of the computational modules with the hardware prototype, which included microfluidic channels and miniaturized spectrophotometers.
The user interface was designed to be intuitive and user-friendly, enabling even non-specialists to operate the device efficiently. It included features for data visualization, real-time result display, and easy navigation of diagnostic functionalities. This required a thorough understanding of Python libraries for GUI development and interfacing with external hardware components.
The project’s broader impact lies in its potential to revolutionize pathogen detection, particularly in resource-limited settings. Unlike conventional methods such as PCR, which require expensive equipment and significant processing time, this platform is designed to be portable, affordable, and user-friendly. Such a tool could be pivotal in addressing outbreaks and monitoring public health, especially in remote or underserved areas.
Conclusion
Working under Professor Kantimay Das Gupta’s mentorship provided me with a profound understanding of interdisciplinary research and problem-solving. The experience honed my skills in computational diagnostics, mathematical modeling, and user-centric design, while also nurturing my ability to collaborate effectively within a research team. This project was a significant milestone in my academic journey, reinforcing my commitment to leveraging science and technology to address real-world challenges.
Internship Certificate

